SnapGene Version 6.1.0
SnapGene 6.1.0 was released on July 04, 2022.
Overview
Version 6.1 simplifies primer design when simulating Golden Gate Assembly, includes a new Secondary Structure view for ssRNA sequences, and supports Dark mode across all operating systems.
Golden Gate Assembly
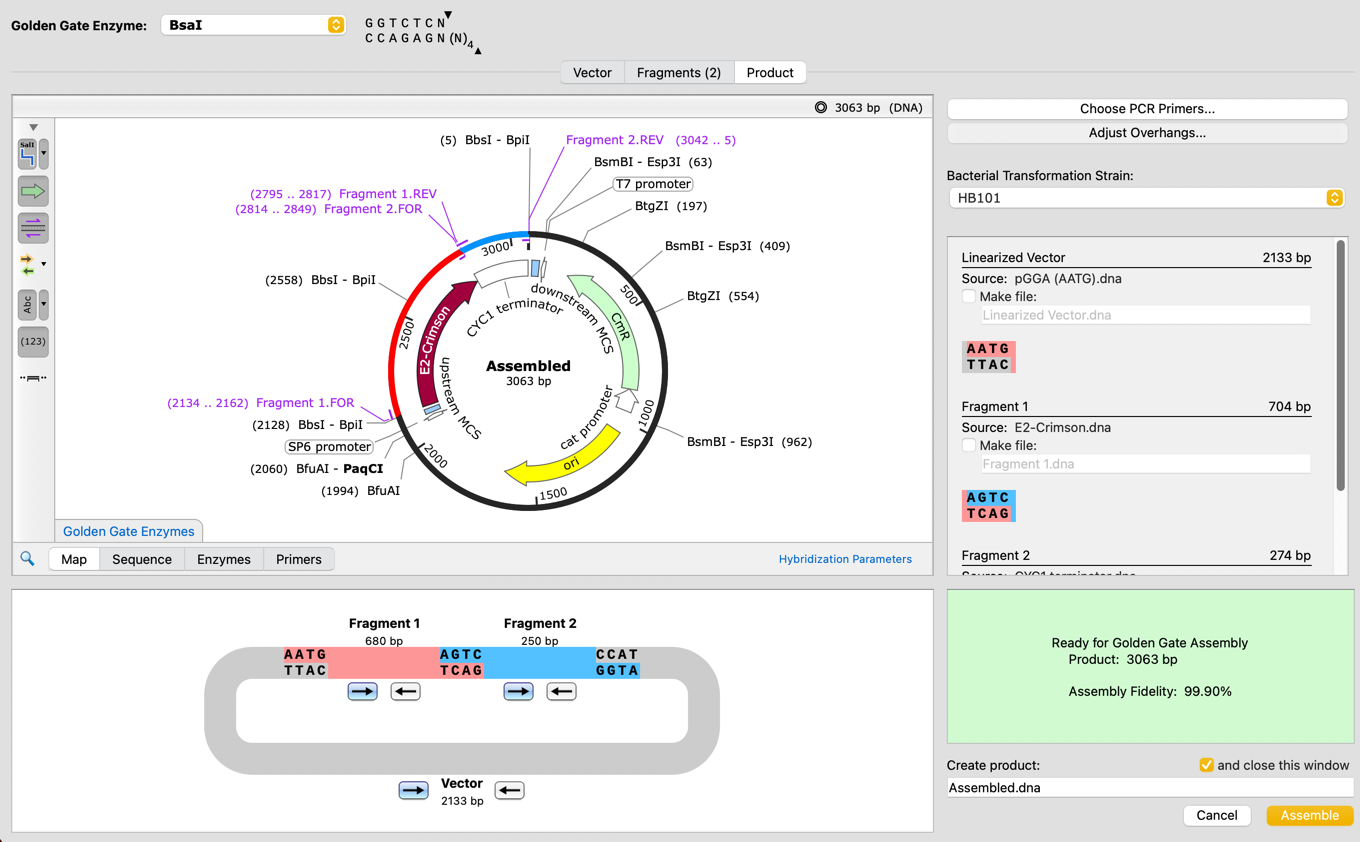
Confidently simulate Golden Gate Assembly with a new tool which can automatically design primers and overhangs for Golden Gate, optimizing the reaction fidelity to maximize the likelihood of success.
RNA Secondary Structure
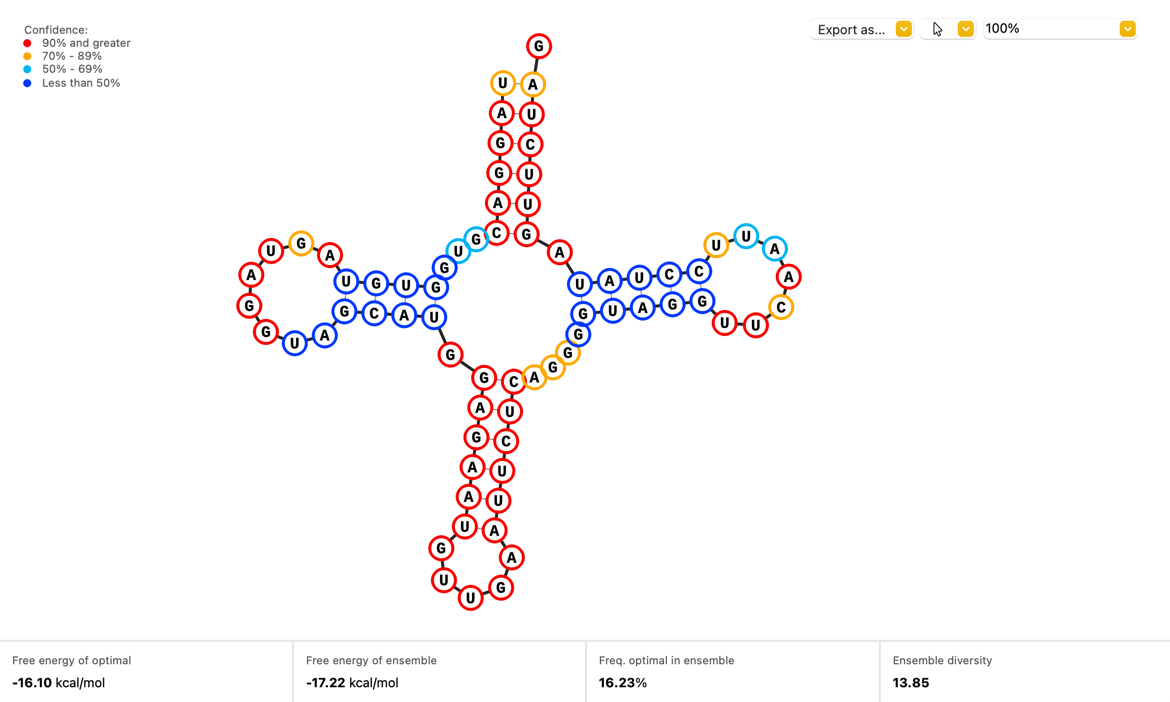
Visualize how your single stranded RNA sequences will fold with a new Secondary Structure view which displays the optimal structure as calculated by the ViennaRNA.
Dark Mode
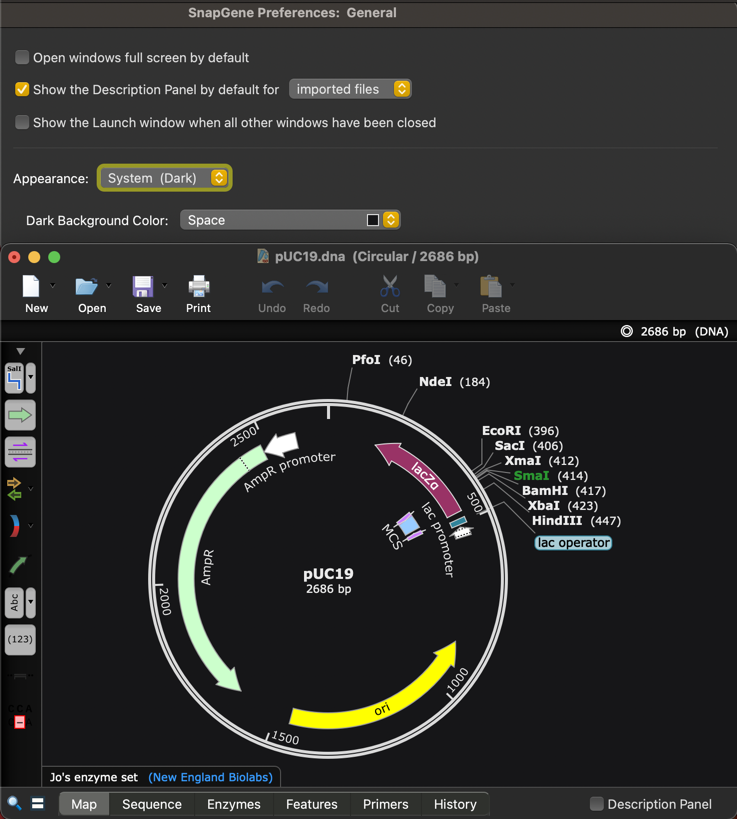
The SnapGene user interface now supports dark mode and by default will match your Dark or Light mode operating system setting.
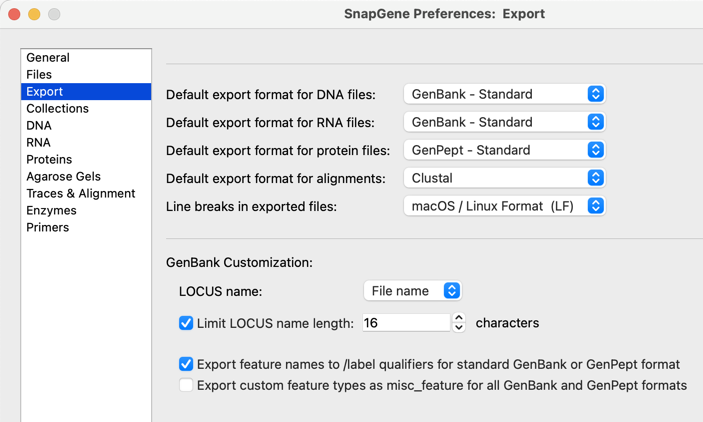
Export Options
Customize how contents is exported to GenBank including LOCUS field identifier feature export options using the new Export pane in Preferences.
Support for Apple Silicon
SnapGene now runs natively on Apple computers with M1 silicon chips.
New Functionality
- New Golden Gate Assembly simulation tool, either using existing Type IIS enzymes or through automated primer and overhang design
- New Secondary Structure view tab for single stranded RNA sequences. Structures calculated using the ViennaRNA package
- Added support for Dark mode, automatically using the OS settings for macOS and Windows
- Native support for Apple Silicon machines
- Enabled customization of content exported to GenBank including LOCUS field identifier and feature export options
Enhancements
- Added "Synthetic Protein" and "Unknown" options for the Source attribute in the description panel
- Added DNA ladders from OriGene
- Ladder fragment lengths are now displayed in the fragment list in agarose gels
- Added LOBSTR to the list of E. coli strains
- Improved the detection and simplified the display of primer hybridizations with 3' overhangs
- Provide a cancel option when a message is shown indicating an unknown or not installed custom feature type must be addressed before proceeding with editing or duplicating one or more features
- Added links to SnapGene Academy
- Extensive optimizations
- Various textual and layout enhancements
Fixes
- Improved stability
- Fixed various memory leaks
- Fixed issues with opening some GenBank files
- Ensured the fragment list in cloning dialogs continue to refresh when either enzyme is manually cleared
- Improved text spacing after punctuation characters on macOS
- Fixed an issue that prevented the Enzymes view "Select Range" context menu command from working
- Improved mouse pointer shape when over links in Collection windows
- Fixed or improved various links to SnapGene.com pages
- Fixed an issue where cloning dialogs could get stuck generating the product
- Fixed an issue where some windows omitted the title bar on Windows
- Fixed an issue where selected codons were not always shown using a white foreground in Sequence view
- Fixed an issue where Ready to Clone was still shown after adding or removing a fragment, even if cloning was no longer possible
- Use the default file format specified in Preferences when exporting alignments
- Fixed an issue with Geneious file import where features in protein sequences could erronously be translated
- Fixed an issue with Geneious file import where introns in nucleotide sequences imported were translated
- Fixed an issue where the Enter key in the numpad was not responsive when a combo box or line edit had focus on macOS
- Improved image quality when exporting to EMF format
- Improved display of checkboxes on Windows
- Fixed dropdown menus in Windows which sometimes had clipped text
- Fixed issues with the display and behaviour of default buttons in dialogs
- Fixed an issue with files appearing disabled in the macOS file explorer when they were able to be chosen
- Fixed an issue that prevented pasting into the Find control in the Open and Save As dialogs on macOS
- Fixed an issue that prevented importing AddGene sequences that contain reserved characters in the file name into a Collection on Windows
- Corrected an issue that resulted in an incredibly wide window when importing a database from CSV format that lacks headers into a Collection
- Fixed an issue where ancestral sequences that cannot be resurrected were shown as blue links in the text format of History view
- Fixed an issue where the Add Primer command could add a primer to an open DNA window even if a Collection window was active when the command was triggered
- Fixed an issue with the display of an amino acid selection highlighting within a translation
- Fixed an issue on Ubuntu whereby AB1 files in a Collection opened in SnapGene Viewer if installed
- Fixed some issues with dialogs opening in strange positions, such as on a different monitor
- Addressed a stability issue when editing a collection description.
- Fixed an issue with importing mixed content types from NCBI
- Fixed various issues with the sequence name when importing from NCBI, especially when using the COMPLEMENT operator.
- Fixed a number of printing issues on Windows when the operating system is set to scale all content to 100% or 150%.
- Fixed the appreance of the transformation strand menu in the Change Methylation dialog on Windows.
- Fixed printing the header for multiple sequence alignments on Windows using 100-200% scaling.
- Improved printing margins on Windows.
- Improved print quality of mutiple sequence alignments on Windows.
- Imporved the default height of the Insert Codons dialog.
- Improved the horizontal alignment of text in the fragmnets list shown in Agarose Gel documents.
- Improved the display of site feature labels in multiple sequence alignemnts.
- Fixed an issue where browsed or edited common features are not realible shown with all relevant qualifiers in the Browse Common Features dialog.
- Repliably respect the "Detect either an exact protein match or approximate DNA match" setting when detecting common features.
- Improved margins when printing on Windows
- Fixed horizontal alignment of enzyme names when printing on Windows.
- Addressed various issues when printing on Windows with the display scaling set to 200% or higher.
- Improved the width of spinboxes on Windows.
- Avoid elliding menu commands on Windows
- Fixed various issues that made it difficult to install software updates on Windows.
- Improved behavior of progress bar when when installing software updates on macOS.