What is Golden Gate Assembly?
Golden Gate assembly, also known as Golden Gate cloning, is a one-pot, one-step cloning procedure created by Carola Engler and colleagues in 2008. The method takes advantage of Type IIS restriction enzymes (e.g. BsaI), which cleave DNA outside their recognition sequences. The result is an ordered assembly of a vector and one or more DNA fragments.
How Does Golden Gate Assembly Work?
Golden Gate assembly can be split into two distinct steps that occur within the same reaction:
- Type IIS restriction enzyme digestion
- DNA ligation
Step 1: Type IIS Restriction Enzyme Digestion
The destination vector and insert fragment(s) both contain compatible Type IIS restriction sites.
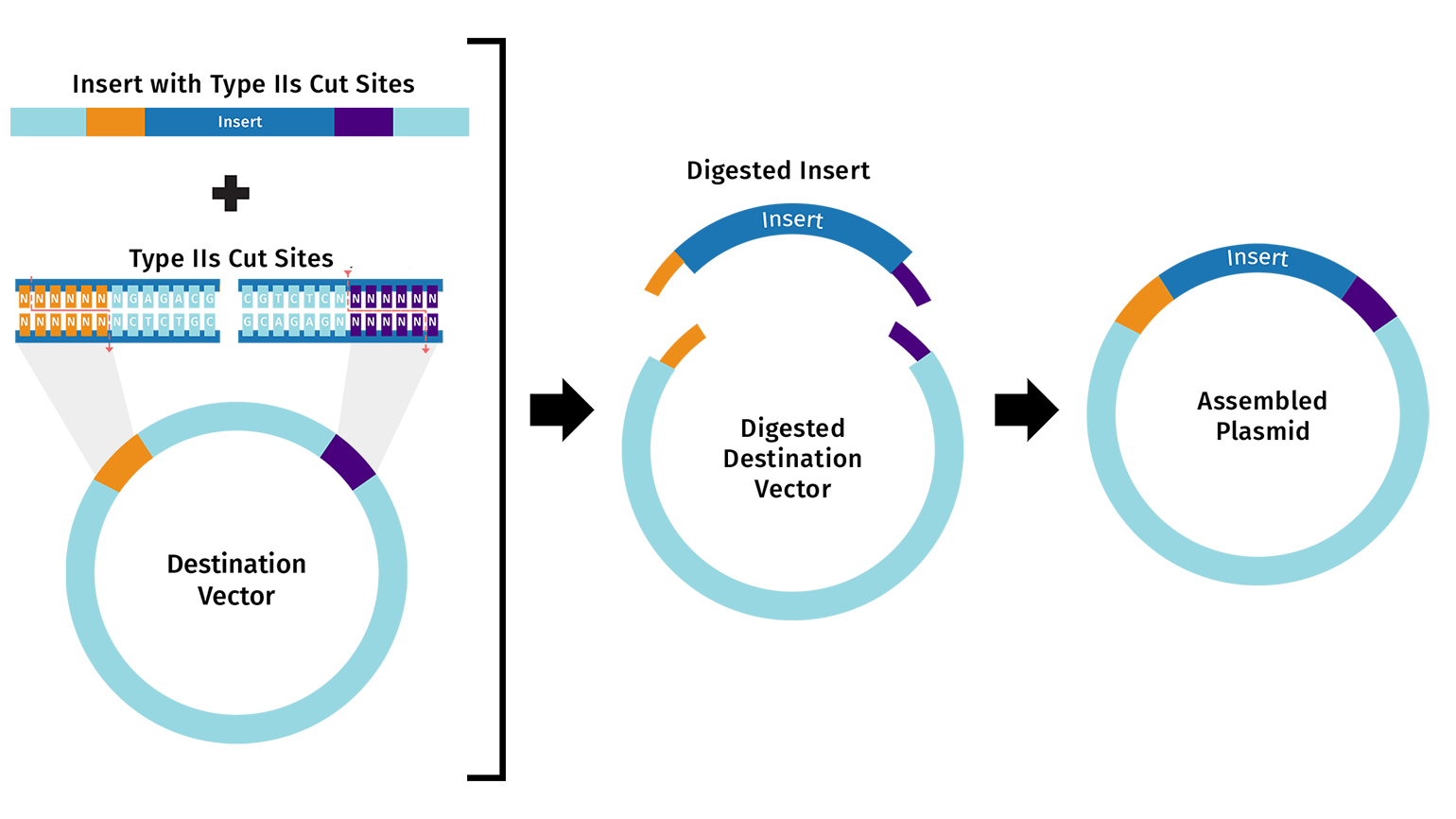
Unlike conventional restriction cloning, which utilizes Type IIP restriction enzymes that recognize a palindromic sequence and cleaves within the recognition site, Golden Gate assembly uses Type IIS restriction enzymes.
Type IIS restriction enzymes have various unique properties that make Golden Gate assembly possible.
- Non-palindromic recognition site: The recognition site is non-palindromic. In general, sites range from 4 to 7 nucleotides
- Shifted cleavage: Cleavage is performed outside the recognition site. The shifted cleavage site allows for sequences to be digested for cloning without disruption of important sequences
- Variable sticky ends: Cleavage on each strand is staggered, resulting in unique overhangs (1 to 5 nucleotides) associated with a single recognition site. For example, with a 4-base overhang, up to 256 different overhang sequences are possible, which enables multiple DNA fragments to be assembled in the same reaction. The unique four base pair overhangs are often referred to as Fusion Sites.
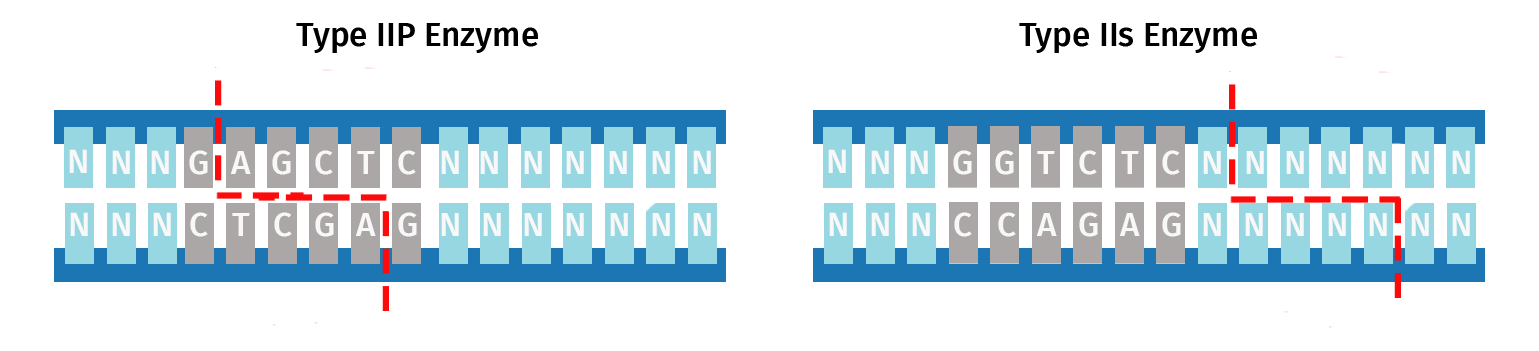
Type IIS restriction enzymes exhibit great diversity in the length of their recognition site, the offset of the cleavage site from the recognition site, and the nature of the overhang; see this list for a complete picture of this diversity.
► Learn more about restriction cloning
Step 2: DNA Ligation
Once the destination vector and DNA insert(s) are digested, their complementary overhangs are joined together by DNA ligase to create an ordered assembly. The process is often denoted as “scarless” or “seamless” since undesired nucleotides are not added between the DNA fragments and the restriction sites are eliminated in the final construct.
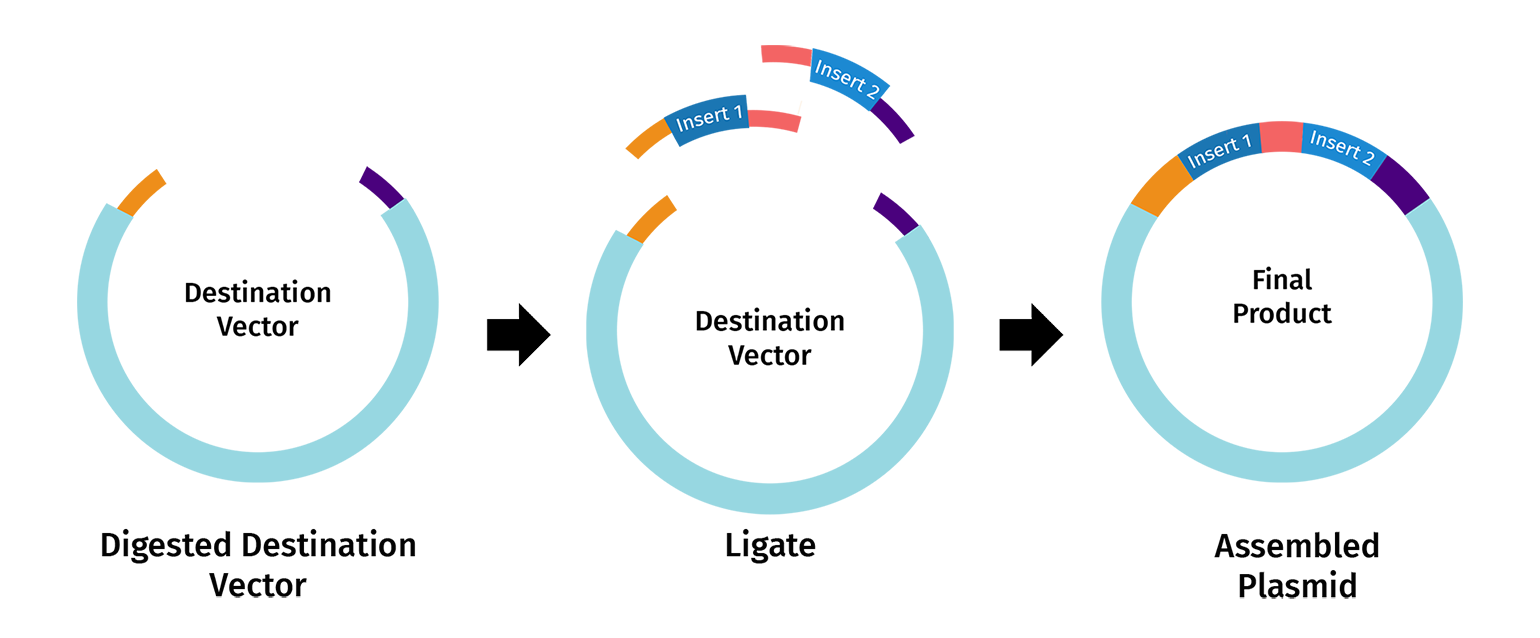
Components of Golden Gate Assembly
Golden Gate assembly works by mixing the following components into a single reaction tube:
- Destination vector
- DNA insert(s) (e.g. amplicon or pre-cloned)
- Type IIS restriction enzyme (e.g. BsaI)
- T4 DNA ligase
- Reaction buffer
Gateway Reactions are processed in a thermocycler. Assemblies of more than 3 fragments will cycle through reaction temperatures to achieve optimal cloning outcomes.
Destination Vector
Golden Gate destination vectors are available either through commercial sites (e.g. New England Biolabs), individual labs, or Addgene.
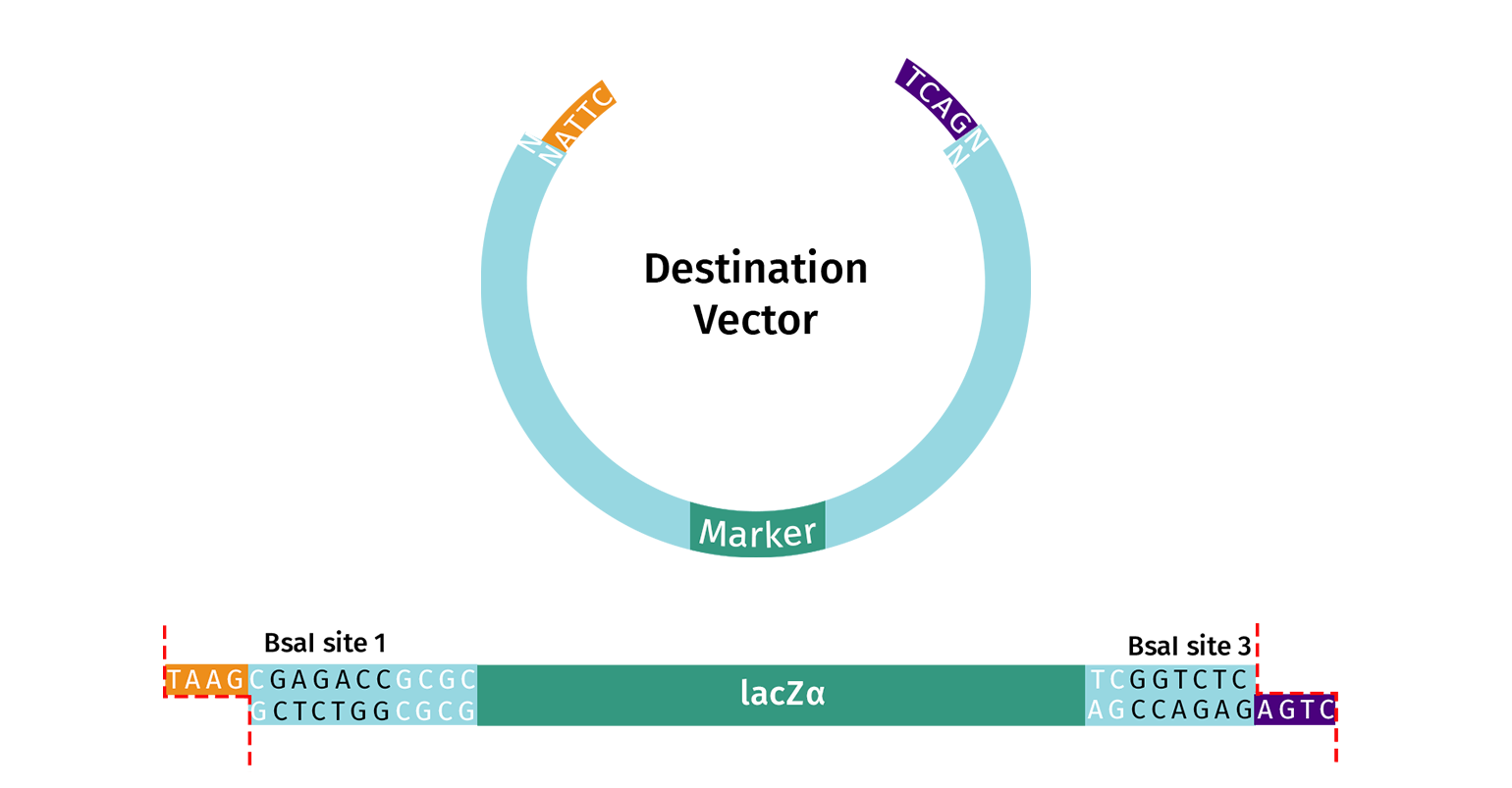
Alternatively, you can adapt your own vector to be Golden Gate ready. The vector should contain two Type IIS recognition sites (e.g. BsaI) that flank the desired insert region and a screening marker (e.g. lacZ). The enzyme recognition sites must be oriented with an “outward” orientation so that after digestion the insert and the enzyme’s recognition sites are removed from the vector.
It’s critical to remove undesired Type IIS cleavage sites from your destination vector; this is known as domestication. If your vector does contain unwanted cleavage sites within the backbone, you will need to remove these before starting Golden Gate assembly. Unwanted cleavage sites can be easily removed with site-directed mutagenesis.
► Watch a video on site-directed mutagenesis
DNA Insert(s)
Any double-stranded piece of DNA can be used as the source of insert for Golden Gate Assembly, including both PCR product and plasmid DNA.
A fragment of interest is adapted for Golden Gate Assembly by the addition of Type IIS restriction sites. Recognition sites must be oriented with an “inward orientation” so that after cleavage the Type IIS recognition site is removed from your fragment.
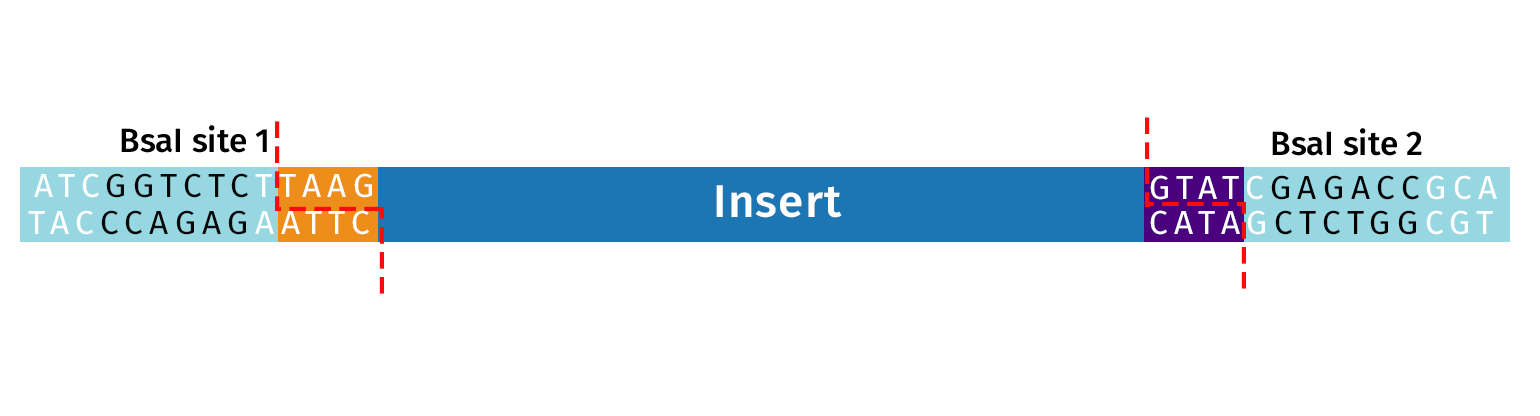
For amplicon preparation, PCR primers are designed that contain flanking bases, the Type IIS recognition site and an overhang sequence. After amplification, the PCR product can either be cloned into a Golden Gate Destination vector or into a Golden Gate Entry Vector, such as pGGA, for sequence verification and re-use in multiple experiments.
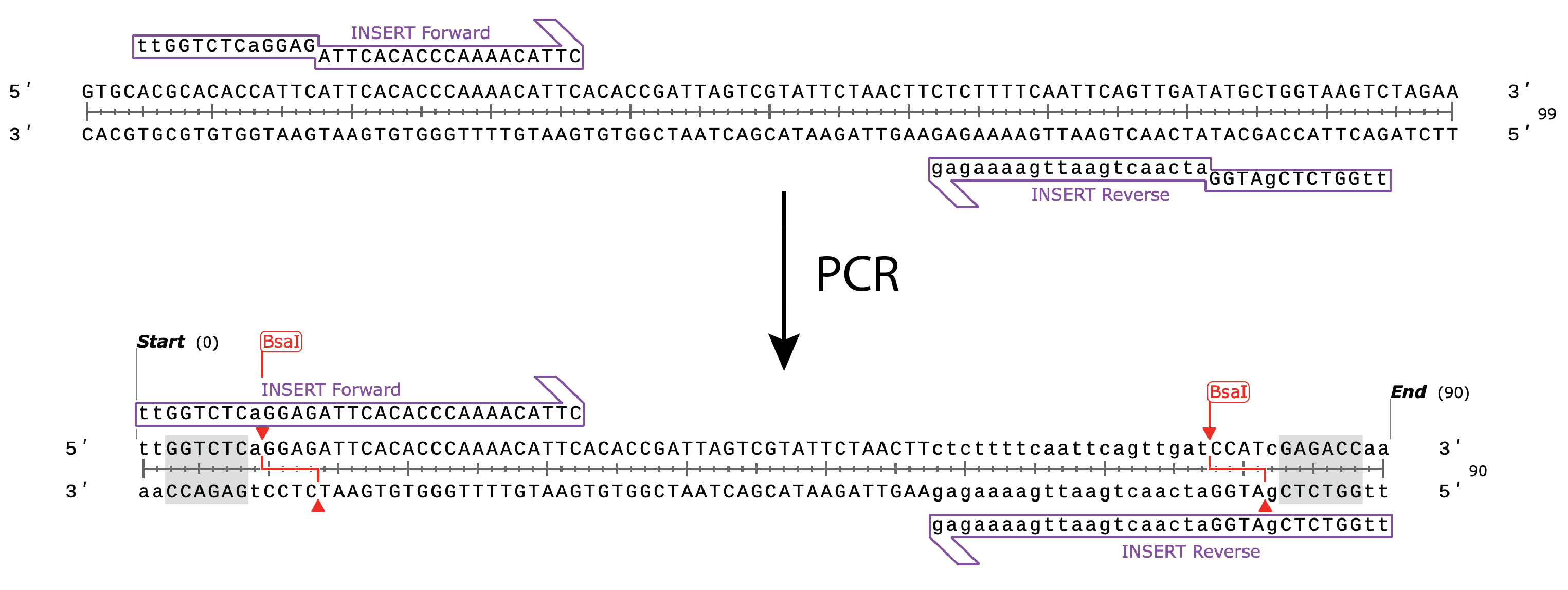
- Forward Primer: 5’ tt GGTCTC a GGAG attcacacccaaaacattc
- Reverse Primer: 5’ tt GGTCTC g ATGG atcaactgaattgaaaagag
As with the destination vector preparation, it’s also important that your DNA insert(s) do not contain extraneous Type IIS restriction sites. Unwanted cleavage sites can be removed with site-directed mutagenesis. If the Type IIS recognition site is in a coding sequence, the mutation(s) you introduce must be silent.
Golden Gate Assembly-Based Kits
There are various pre-designed kits available that utilize Golden Gate technology, such as transcription activator-like effector nucleases (TALENs), Golden GATEway and modular cloning (MoClo).
TALENs
Thomas Cermak and colleagues first described the design and assembly of custom TALEN constructs in 2011. TALENs are fusions of transcription activator-like (TAL) effectors to a Type IIS restriction endonuclease (e.g. FokI), which is achieved through Golden Gate assembly. TAL effectors contain a customizable array of amino acid repeats that bind to desired regions of DNA. Once bound to DNA, the fused nuclease creates a targeted double-strand break. An alternative to CRISPR, TALENs are useful genome engineering tools that can be used to create gene knockouts or introduce specific DNA sequence modifications.
Golden GATEway Cloning
Golden GATEway cloning kits combine the efficiency of Golden Gate assembly with the sequence-independent approach of Multisite Gateway™ cloning.
As described by Stephan Kirchmaier and colleagues in PLOS One, fragments of interest are cloned into one of eight Golden Gate entry vectors. The elements in the GG-entry vectors are then assembled in one of three Golden Gate destination vectors. The Golden Gate destination clones play a dual role and function as entry clones for three-part Multisite Gateway cloning.
► Learn more about Gateway™ cloning
MoClo
The MoClo system is a hierarchical approach introduced by Ernst Weber and colleagues that enables the creation of multigene constructs through a series of three Golden Gate assembly reactions.
MoClo kits generally contain three sets of cloning vectors (Level 0, 1, and 2) that allow users to recombine basic modules (e.g. promoters, coding sequences, and terminators) into transcription units, which are later joined to form multigene constructs. The MoClo system can be readily automated for applications such as gene stacking and metabolic engineering.
Tips and Additional Resources
Double-check the orientation and order of your fragments
When performing Golden Gate assembly, it’s very important you take the time beforehand to plan the orientation and order of your DNA fragments.
It’s recommended to map the Type IIS restriction sites that you will be using and carefully annotate the fusion sequence associated with each site. When using primers to introduce Type IIS restriction sites to amplicons or pre-cloned plasmids, ensure the recognition sites face inwards (i.e. towards your DNA insert). As you process your cloning strategy, you will need to be sure that the selected fusion sites will ultimately assemble your fragments of interest in the correct order.
Consider different Type IIS restriction enzymes
If your vector backbone or insert fragments contain unwanted Type IIS restriction sites, then also consider using an alternative enzyme. It’s more likely to discover internal sites with Type IIS enzymes that have a shorter recognition site (e.g. MnII, 4 bases), compared with those with a longer recognition site (e.g. BaeI, 7 bases).
Commercially available Golden Gate assembly components
Learn More About Cloning





