The Industry’s Most Popular Molecular Cloning Tool

The easiest way to plan, visualize and document your everyday molecular cloning procedures
Improve accuracy while moving faster, so you save time and money.
- Elegant, information-rich windows for simulating common cloning and PCR methods
- Clear visual schematics let you see exactly how your construct will be put together
- SnapGene helps you identify and avoid common mis-steps by keeping track of details like DNA methylation and phosphorylation
University of Glasgow
Beyond the Basics of Molecular Cloning
SnapGene is the most popular cloning tool for a reason. It’s fast, smart and extremely user-friendly
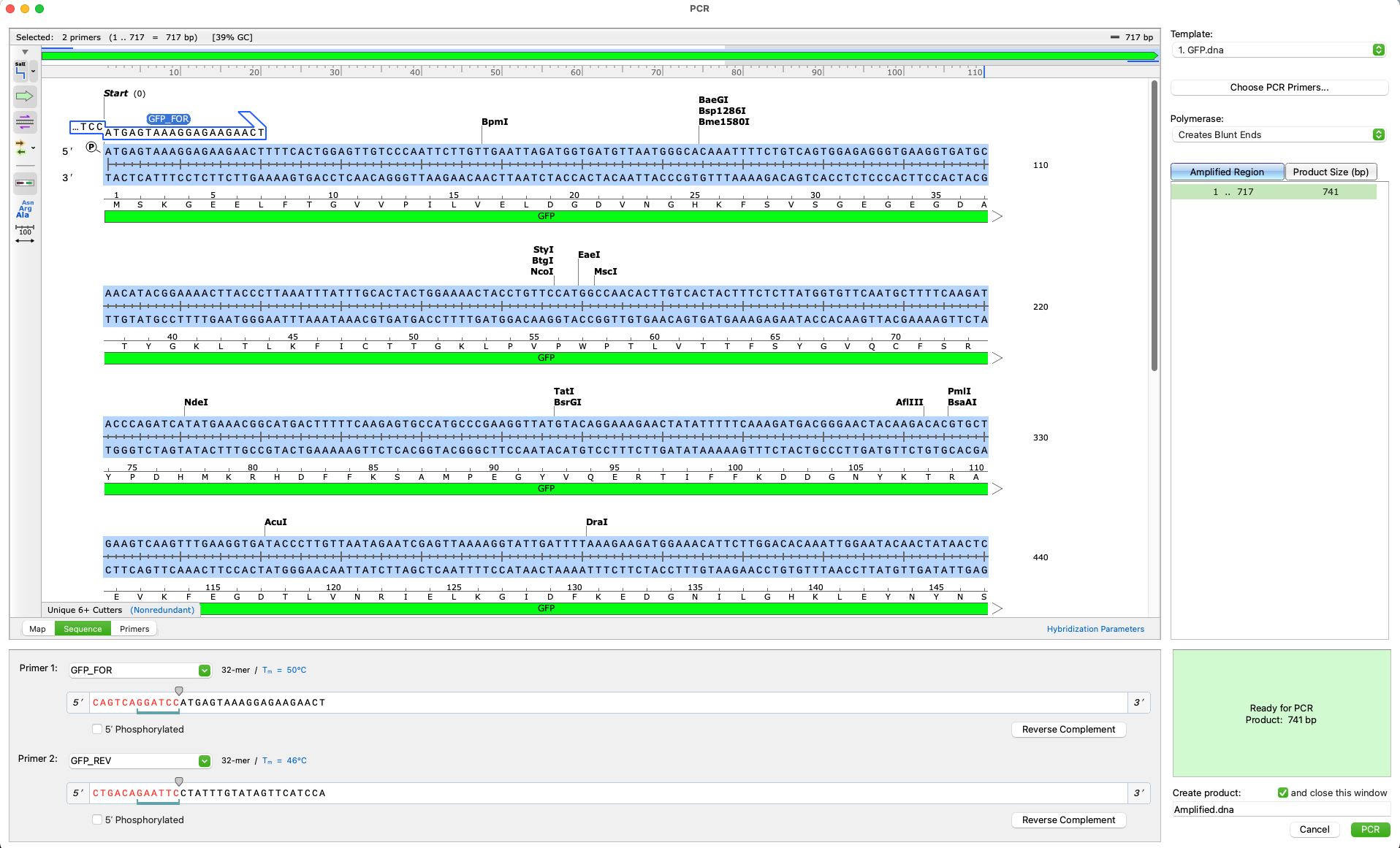
- Intuitive technology identifies design flaws in cloning procedures so they can be corrected
- Simulate standard PCR using your own primers, or allow SnapGene to design them automatically
- Specialised cloning tools ensure fast accurate construct design for all major molecular cloning techniques
Villanova University
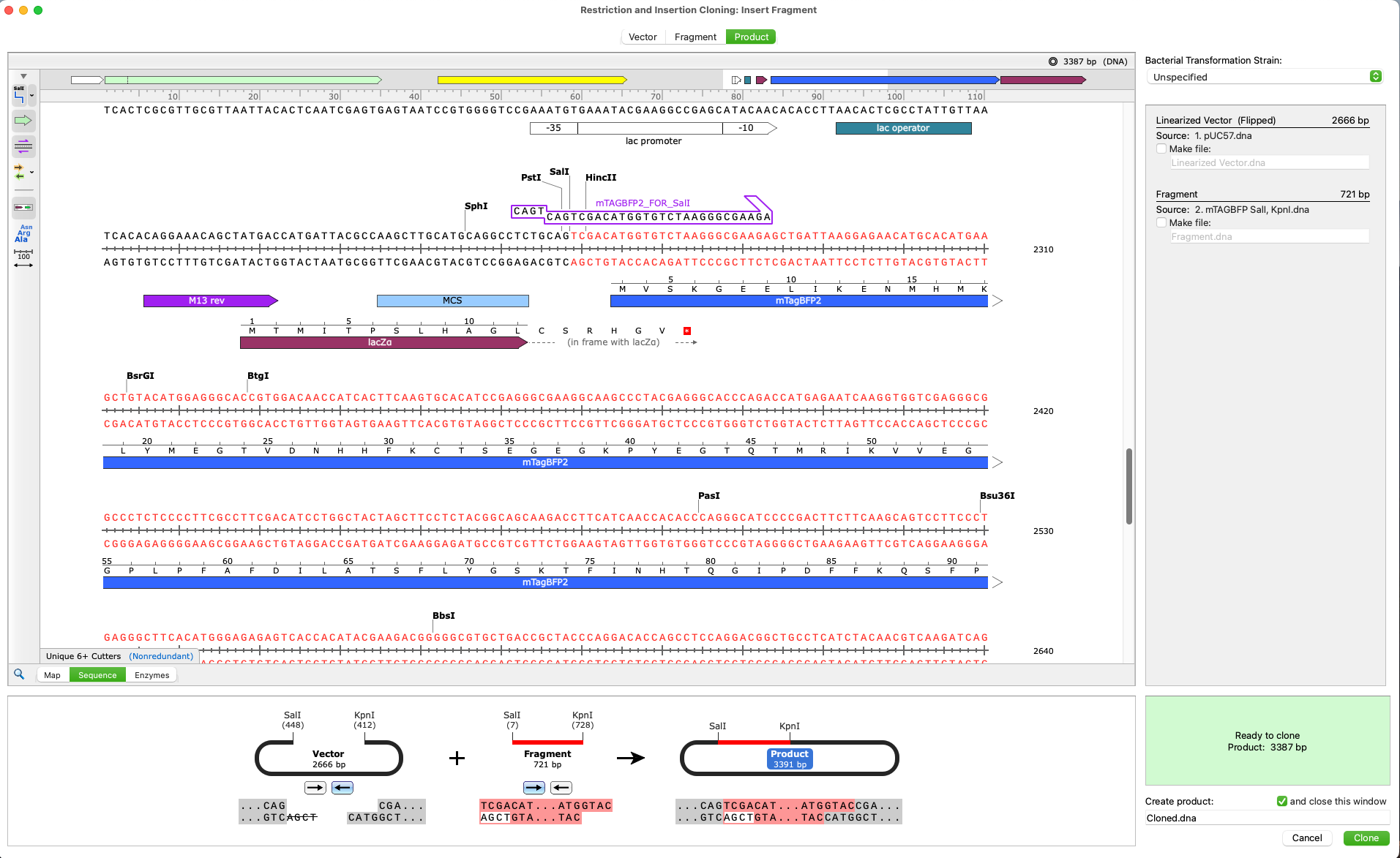
Verify your project design and prevent errors
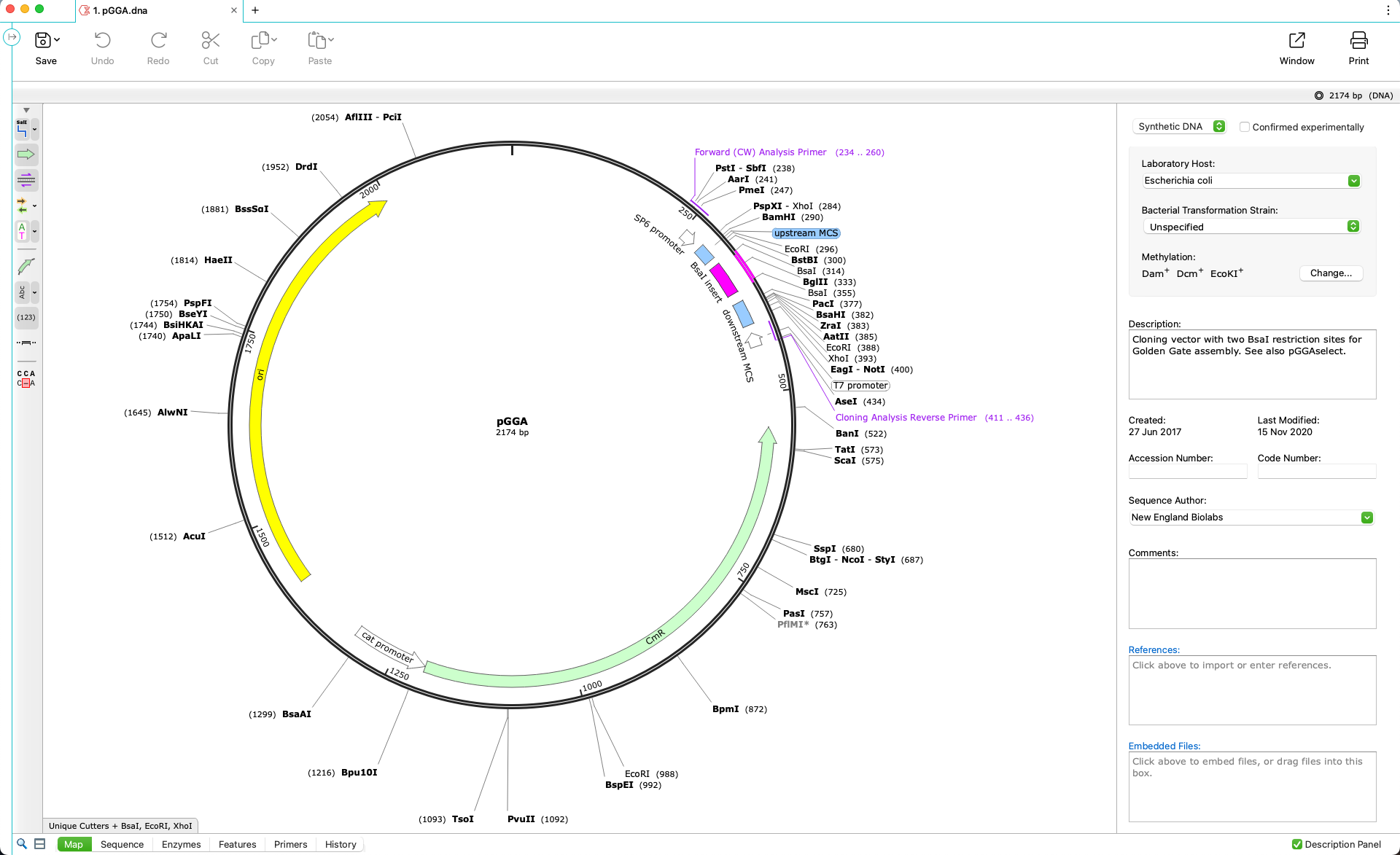
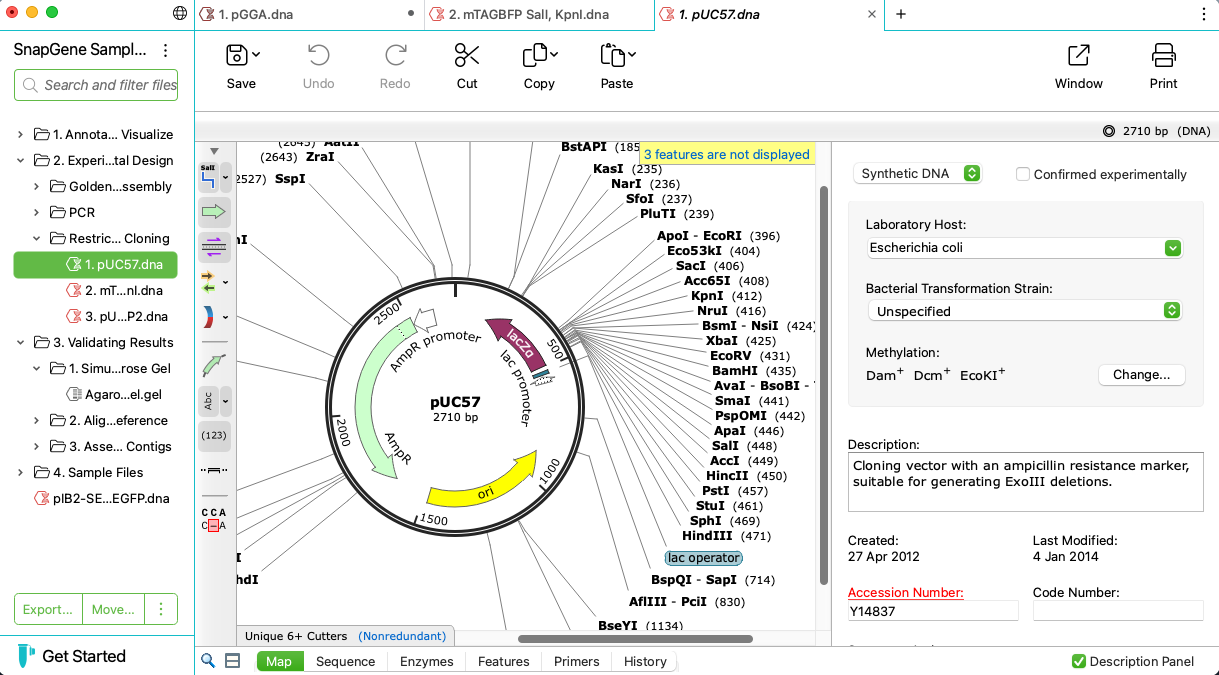
Automatically View Plasmid Features
- Annotate features on your plasmids using SnapGene’s curated feature database or your own custom features
- Display enzyme sites, features, primers, ORFs, translations and more on plasmid maps or in detail on the sequence view
- Customize your maps with flexible annotation and visualization controls
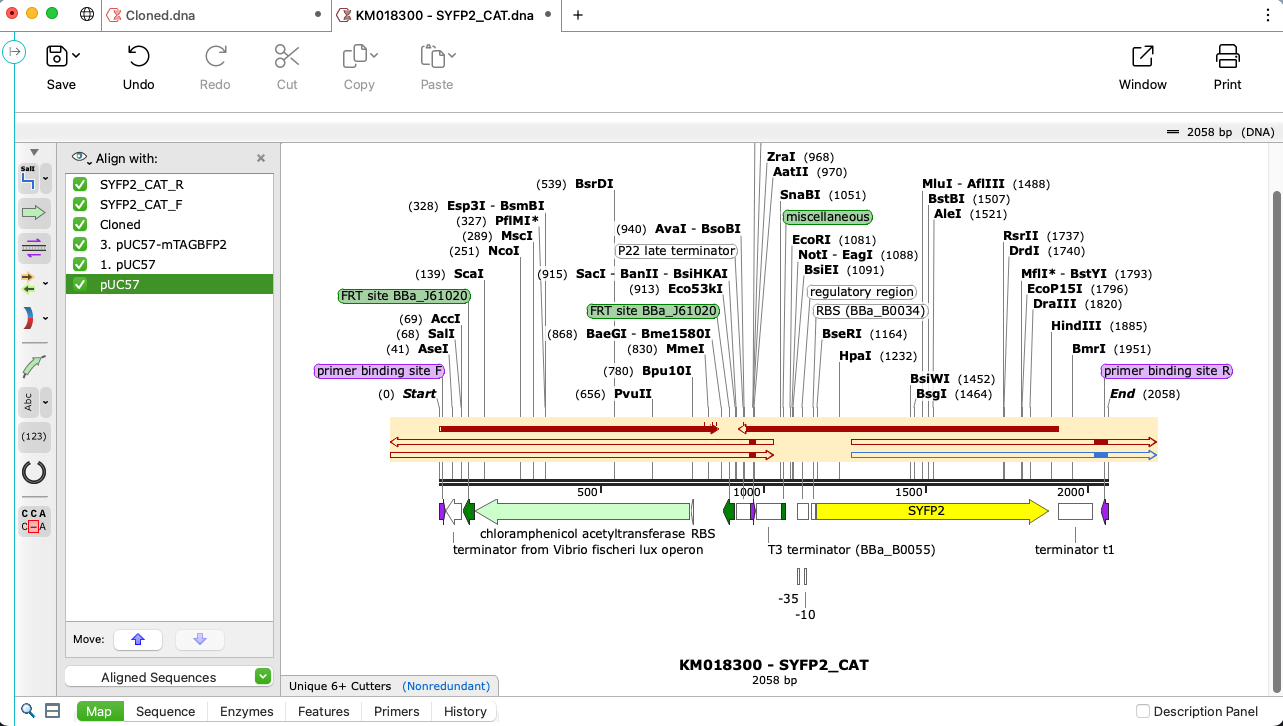
Check Your Sequences with Alignment Tools
- Validate your sequenced constructs match your simulated construct using the powerful Align to Reference tool
- Align sequences using trusted algorithms for pairwise and multiple alignments, including Clustal Omega, MAFFT, MUSCLE and T-Coffee
- Assemble Sanger sequencing reads into complete contigs using CAP3
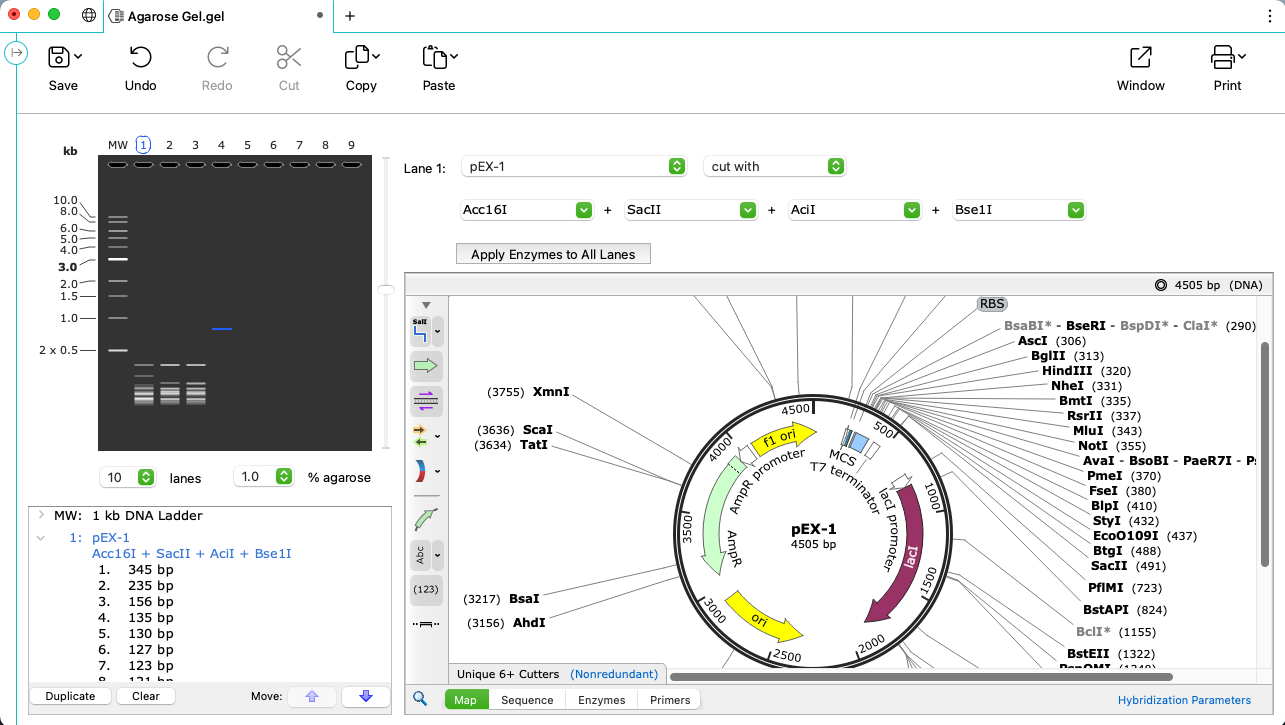
Realistic Agarose Gel Simulations
- Visualise exactly what you will see in the lab with SnapGene’s empirically based gel simulation algorithm
- Flexible configuration of all gel elements, including number of lanes, % agarose, running time and a full set of MW markers
- Record and identify your band of interest with detailed fragment information for each lane
UT Health Science Center, San Antonio
Configured for automatic documentation and easy data exchange
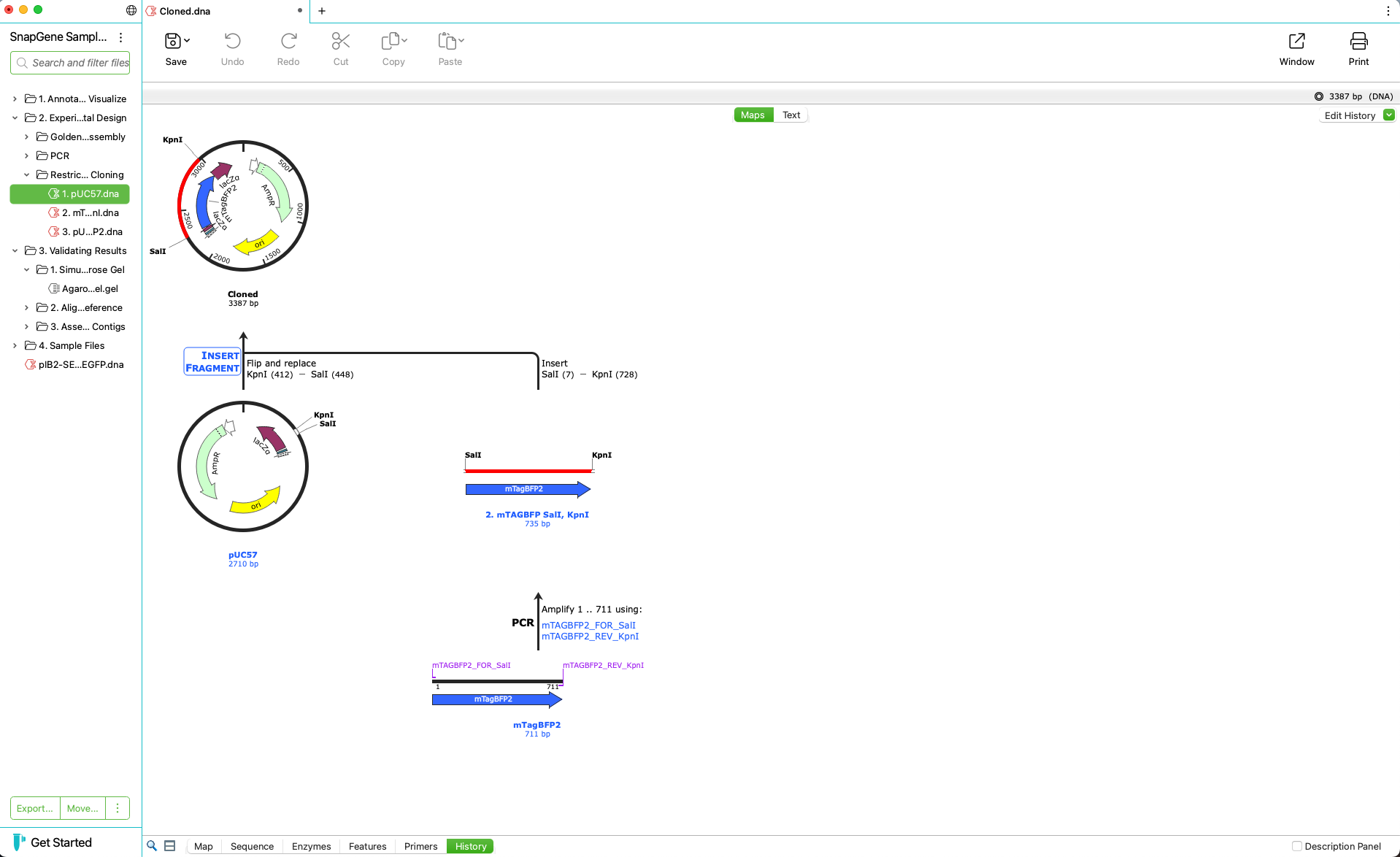
Automatically Create a Graphical History
- Record experimental procedures and changes to documents visually
- Show history colors in history, map and sequence views
- Fully resurrectable sequence history embedded in the document
Convert and Share Data
- Store, search, share and organize your sequences, files and maps
- Work collaboratively with sequences stored on sharable drives, servers or cloud services
- Easily read and export sequences, annotations and other metadata from many common file formats
Fred Hutchinson Cancer Center
All Features
Explore a list of SnapGene’s robust set of features designed to enhance your everyday
molecular cloning procedures
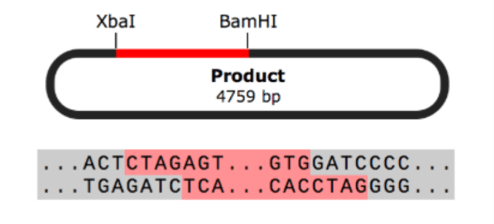
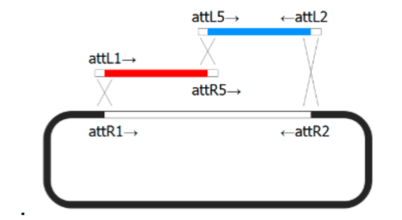
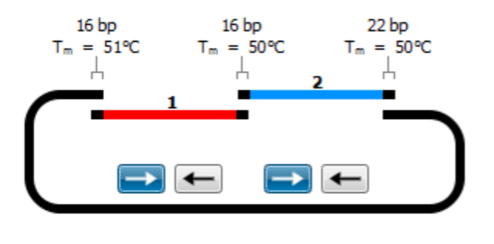
Molecular Cloning
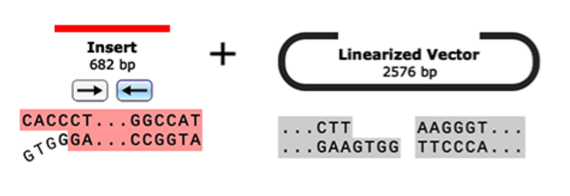
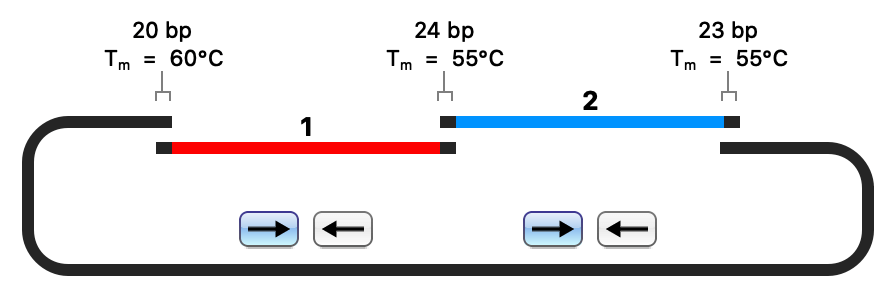
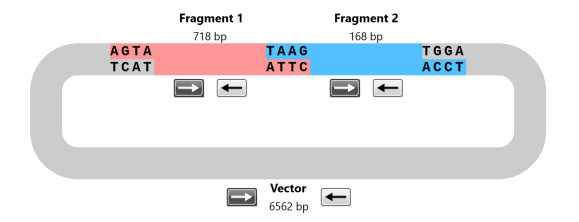
- Restriction Cloning
- Gateway Cloning
- Gibson Assembly
- NEBuilder HiFi Assembly
- In-Fusion Cloning
- TA & GC Cloning
- TOPO Cloning
Primers
- Design primers
- Anneal two oligos to form a double-stranded product
PCR and Mutagenesis
- Simulate PCR
- Overlap extension PCR
- Primer-directed mutagenesis
Enzyme Sets
- Predefined enzyme sets - by company or cutter
- Create custom enzyme sets
- View detailed enzyme information
- Rich support for methylation sensitivity and associated error prevention
Convert File Formats
- Alignment Formats
- ApE
- CLC Bio
- Clone Manager
- DNA Strider
- DNADynamo
- DNASIS
- DNAssist
- DNASTAR Lasergene®
- DS Gene
- EMBL (ENA)
- EnzymeX
- GenBank / DDBJ
- Gene Construction Kit®
- Geneious
- GeneTool
- Genome Compiler
- Jellyfish
- MacVector
- pDRAW32
- Serial Cloner
- Swiss-Prot
- Vector NTI®
- Visual Cloning
Agarose Gel Simulation
- Simulate an agarose gel
- Simulate a Restriction Digest
- Simulate a PCR Amplification
- Large collection of MW markers
Features / Annotations
- Create and edit features
- Automatic annotation of common features
- Annotate novel features manually
- Choose Alternative Codons
- Sophisticated numbering of feature translations
- Support for ribosomal slippage
Translations
- View and edit translated features
- Open reading frames (ORFs)
- Whole-sequence translations
- Check reading frames for gene fusions
- Make Protein (from DNA)
- Reverse Translate (from Protein)
Alignment
- Align DNA sequences with a reference sequence
- Verify cloning or mutagenesis
- Align cDNA to a chromosome
- Pairwise and multi sequence DNA and Protein alignment
- Choice of alignment algorithms - Clustal Omega, MAFFT, MUSCLE, T-Coffee
- Contig Assembly
Visualizing
- See multiple views of a DNA sequence
- Large sequence support - browse chromosome size sequences
- Edit DNA and protein sequences
- Color code sequences
History Tracking
- Comprehensive “undo” capability
- See a graphical history of a product
- Use optional history colors to identify the most recent change to a sequence
Data Management
- Import from common file formats including annotations and notes
- Export to standard formats
- Create and share Collections
- Share data with SnapGene Viewer
- Run batch operations
Search
- Search for DNA or protein sequences
- Search for enzymes, features or primers
General
- Cross platform compatibility - Windows, macOS, Linux
Discover the most user-friendly molecular biology experience.