SnapGene Version 7.1.0
SnapGene 7.1.0 was released on November 28, 2023.
Overview
SnapGene 7.1 provides enhancements to streamline file search within a selected folder, introduces secondary structure visualizations for ssDNA sequences including primers, and includes a variety of fixes to improve stability, file import workflows, and other functions.
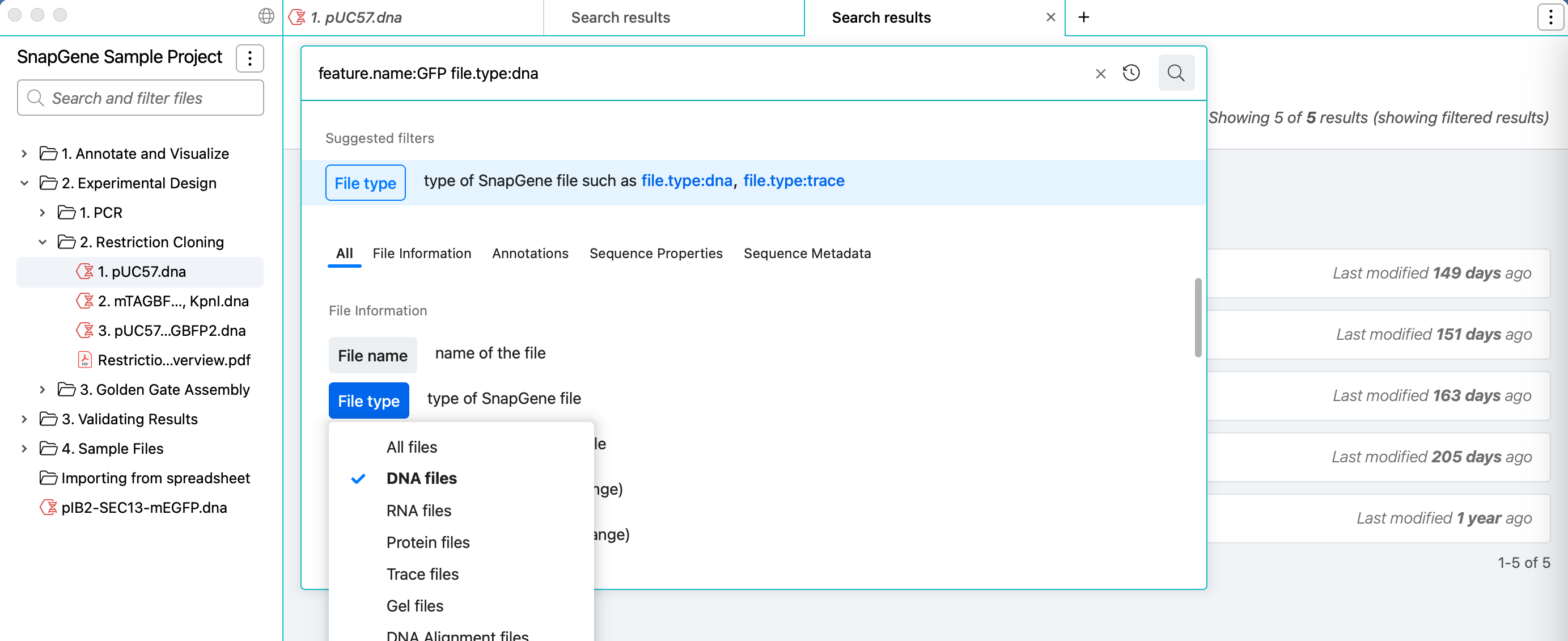
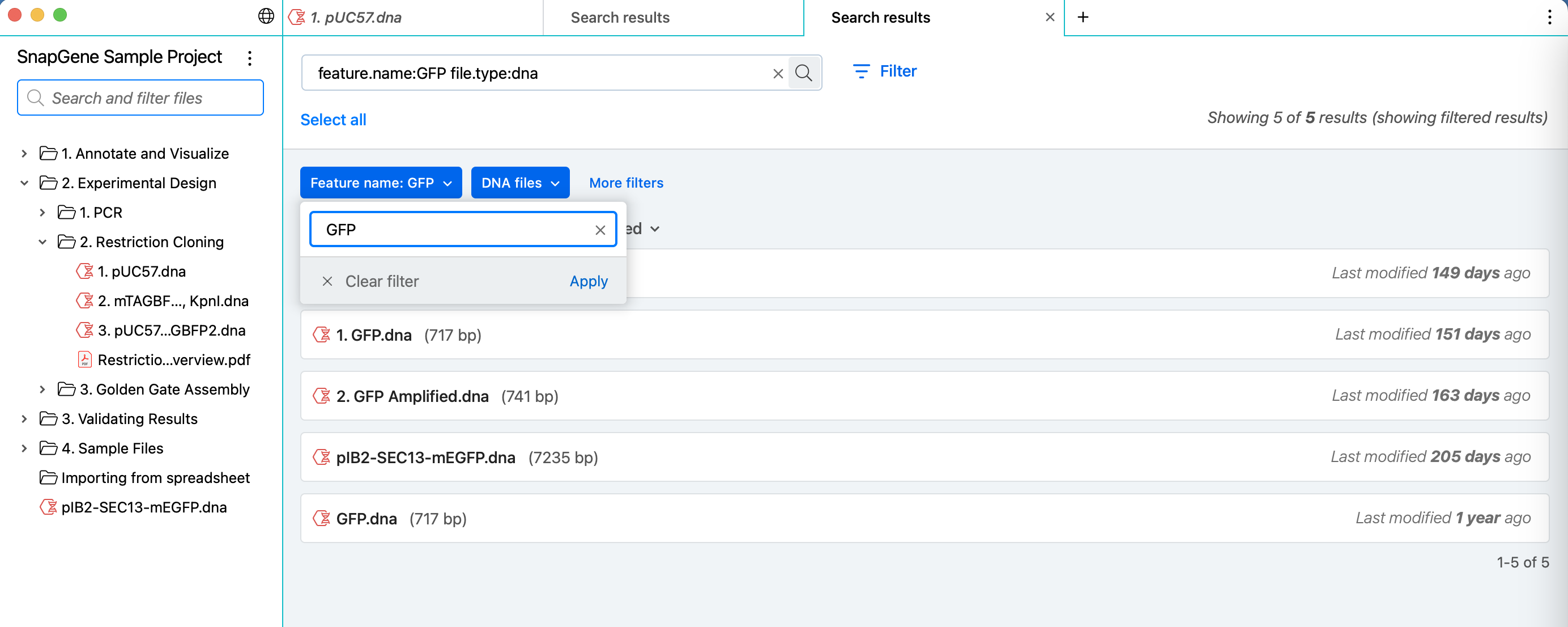
File Search Enhancements
Quickly and easily build search queries to find your sequences and other files in the selected project folder with simplified filter entry controls, and run multiple searches in different tabs.
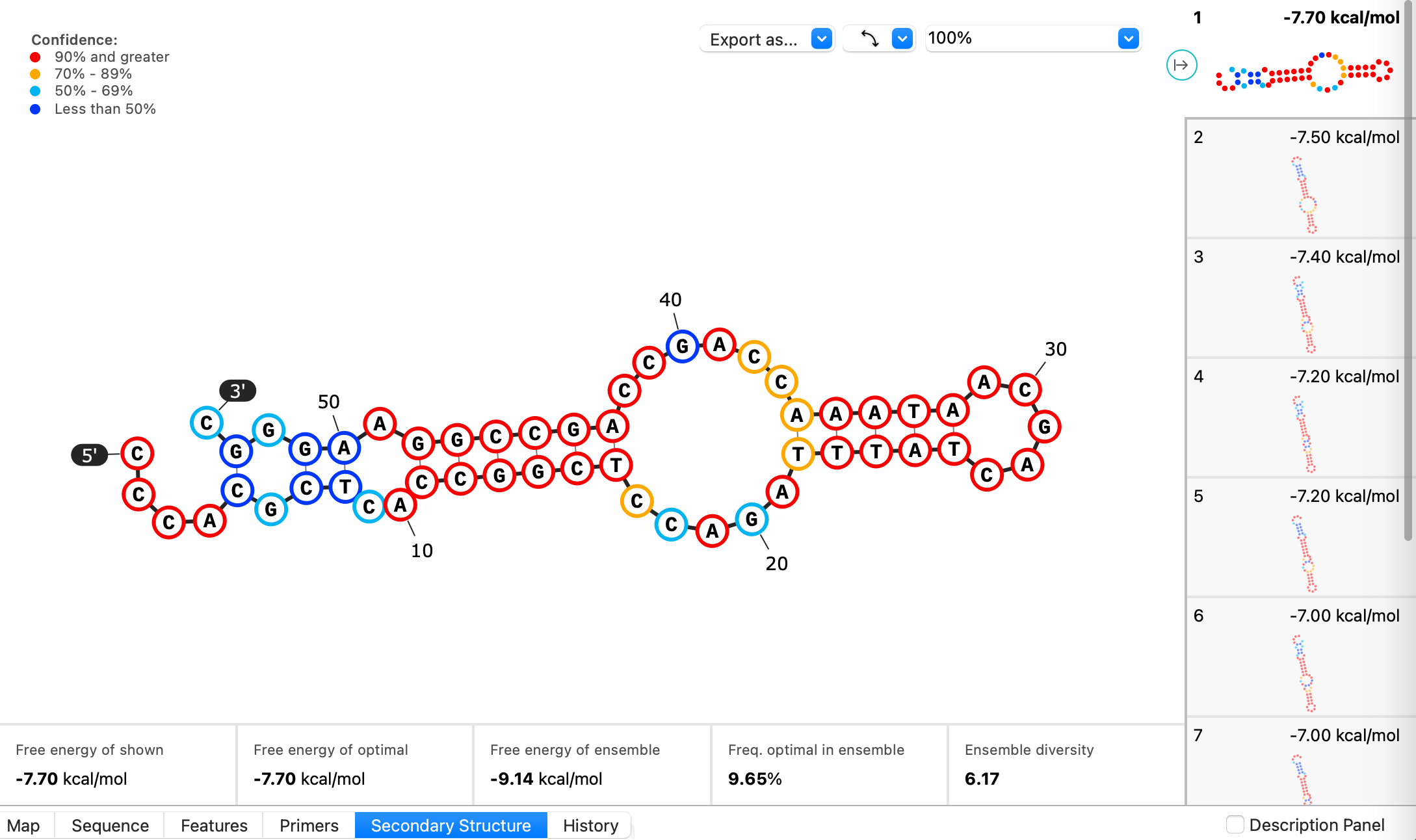
Secondary Structure View for Single-Stranded DNA Sequences
Visualize the secondary structure for single-stranded DNA sequences calculated with the ViennaRNA package via the new secondary structure tab
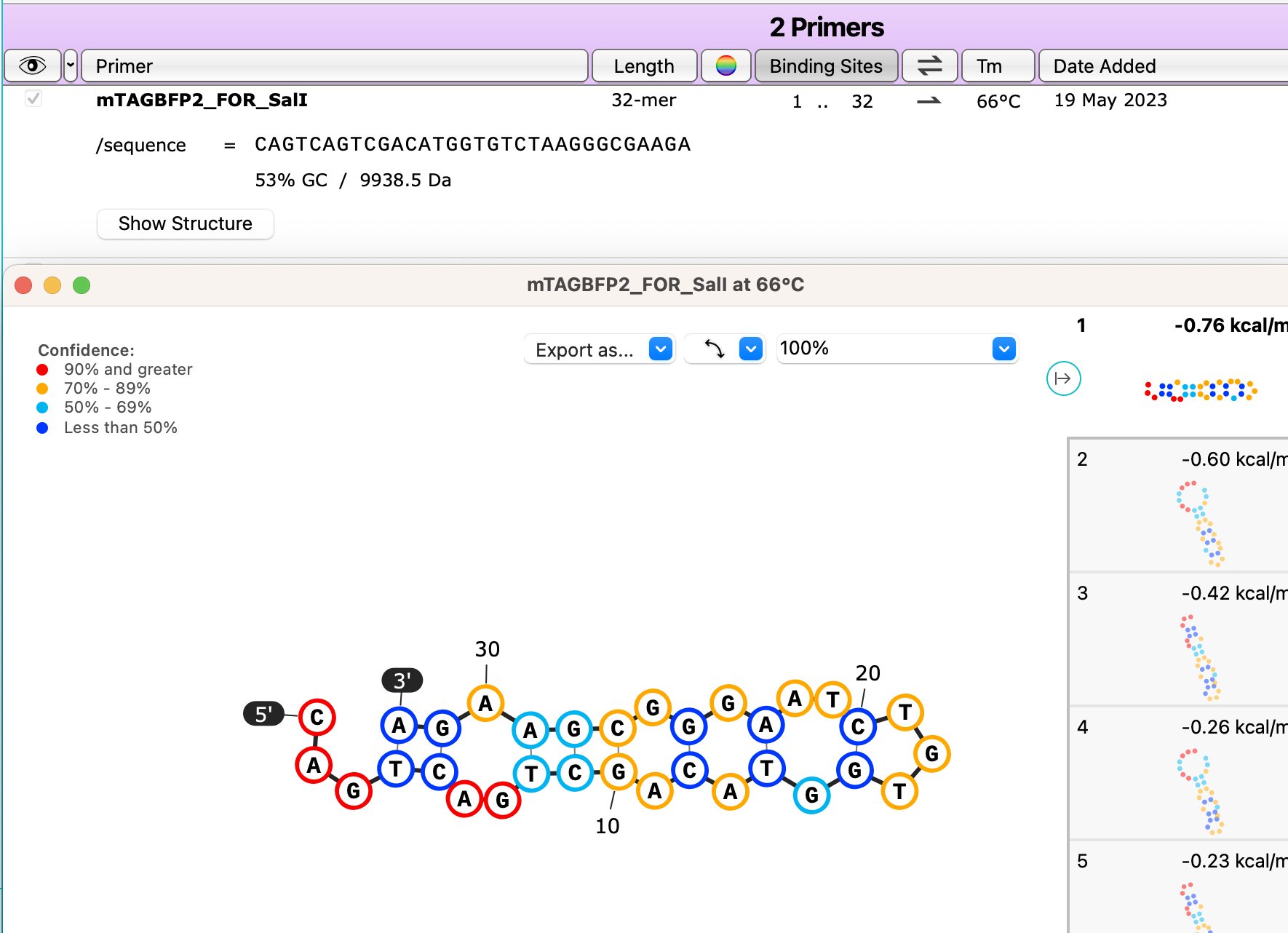
View Primer Hairpins and Secondary Structures
Check your primers for hairpins or other self-binding secondary structures with a new button for calculating secondary structures for primer monomers, available in the primers tab.
Features and Enhancements Include
- Enhanced File Search input filters to streamline and simplify building search queries
- Simple input fields for search terms add search keys automatically
- Date and file type fields provide a dropdown menu for quick selection
- Searching occurs automatically when adjusting the query in a results tab
- File Search results enhancements
- Search results are now shown in a new tab, allowing you to revisit prior search results
- Enhanced visibility and arrangement of active search filters in search results
- Added a secondary structure tab for single-stranded DNA sequences. Secondary structures are calculated using the ViennaRNA package
- Added a “Show Structure” button to Primers view, allowing you to view primer secondary structures
- Structures calculated using the ViennaRNA package
- Primer binding site melting temperatures are used as the default temperature when predicting the secondary structure
- Added a new preference to specify if files that lack a FASTA comment line (a line that begins with “>” before the first sequence) will be recognized and imported as FASTA sequence files, or as text
- The ⌘+W (macOS) / Ctrl+W (Windows/Linux) keyboard shortcuts can now be used to close the Window when viewing the Get Started space
- Restored expanded/collapsed state of the Get Started, Cloning tutorials, and User guide sections on the Get Started space.
Additional Changes and Fixes
- Adjusted handling of KEYWORDS when importing and exporting to GenBank
- KEYWORDS are no longer used as a custom map label when importing GenBank files
- Custom map labels are now exported using a comment
- The alignment dialog no longer uses the custom map label as the sequence name when importing multi-sequence GenBank files
- Adjusted exporting features with segments to GenBank
- Non-gap feature segments are now exported in GenBank files using the join(...) qualifier, including both overlapping segments and immediately adjacent segments
- Fixed an issue where the translation of features with ribosomal slippage (overlapping segments) would change when exporting to and re-importing from GenBank
- Added the /ribosomal_slippage qualifier to any translated features with overlapping segments
- Moved the “Close All Tabs” and “Close Saved Tabs” options to the top of the tab actions menu to ensure they are readily accessible
- Adjust wording of preference to open files as standalone windows by default
- Improved colors used for folders in the Collections left panel to make selected folder names easier to read
- Added a “Close All Tabs” option to the File menu
- Renamed the “Close All” option in the File menu to “Close All Windows”
- Renamed the “Close” option in the File menu to “Close Window” or “Close Tab” as appropriate
- Updated links to SnapGene Academy
- Modified search filter chip appearance to improve contrast
- Improved performance when opening GenBank files by using default colors by feature type when no feature color information is provided.
- Improved handling when entering primer names or selecting primers in cloning and PCR dialogs
- Fixed the Find Enzyme / Feature / Primer shortcut (Ctrl+Shift+F) on Windows
- Fixed an issue with Align to Reference DNA Sequence whereby poor quality reads caused other reads to align incorrectly
- Fixed a bug that resulted in extra bases being deleted from sequence reads when trimming when aligning to a reference DNA sequence
- Fixed the SwissProt importer to correctly read protein sequence names
- Correctly preserve aligned sequence names when importing from multi-sequence archive files
- Fixed an issue with displaying GC colors on some circular sequences
- Prevented automatically rechecking the transfer features checkbox whenever changing a sequence name in the New File pane
- Fixed an issue with converting sequences to circular/linear when the sequence had unsaved methylation changes
- Fixed a bug that resulted in some files being opened using a text viewer instead of being imported
- Fixed bug where a Collection would open in a content window when opening an "Open This Collection.clxn" file
- Create and open a new project instead of a Collection when importing files that contain multiple sequences
- Fixed various issues that resulted in nonsensical warnings being shown when using the Gateway cloning dialogs
- Fixed various issues with searching projects that contain certain non-sequence file types on Linux
- Fixed bug where sequences converted to double-stranded would revert to single-stranded on edit and vice versa
- Fixed a bug where non-sticky tabs weren't marked as "modified" when a document was replaced
- Removed prompt to update older documents when viewing files in a non-sticky tab
- Fixed an issue that prevented unmounting external volumes on Windows that were not in use by SnapGene
- Improved stability and improved performance when compressing data over 2 GB
- Fixed a crash that could occur when using Close All Windows with multiple open tabs
- Improved stability, performance, and addressed various memory leaks
- Added optional anonymous usage reporting to support ongoing development and user research. Note that Personal Information and the contents of the data that you process are not shared.
- Ensure newly created sequences are opened in a new window if the setting is checked
- Improve explanation of how stop codons are treated when performing protein searches
in the Search Tips dialog - Fixed an issue where the Add Primer dialog sometimes thought no template was specified
- Ensure Windows version is properly identified
- Fixed an issue that prevented undoing the removal of 5' overhangs
- Fixed an issue where alignment to a reference was turned off after editing DNA ends
- Fixed a crash that could occur when opening trace files
- Prevent displaying a downstream terminal phosphate after linearizing ssDNA
- Streamlined the Dock (macOS) and Quick launch (Windows) menus
